Introduction to CLIP Sequencing
RNA binding protein (RBP) is a key protein that regulates gene expression at the post-transcriptional level. RBP forms ribonucleoprotein complexes by binding to transcribed RNA. The complex can control RNA biogenesis, stability, cellular localization, and transport, as well as RNA molecules' fate and function. As a result, understanding post-transcriptional regulation in various biological processes requires a high-resolution and accurate protein-RNA interaction map. CLIP is an antibody-based technology used to investigate RNA-protein interactions in RNA immunoprecipitation. The technology uses specific antibodies of RNA-binding proteins to precipitate RNA-protein complexes and retrieve RNA fragments for high-throughput sequencing, uncovering in-depth the regulatory effects of RNA-binding proteins and RNA molecules and their significance to life. CLIP-seq is progressively being used to decode protein complex-mediated post-transcriptional regulation because it can provide a genome-wide map of protein-RNA interactions. Meanwhile, the method can reveal the role of RNA in a variety of biological processes and is a ground-breaking technology that exposes the interaction between RNA molecules and RNA binding proteins across the entire genome.
Advantages of CLIP-Seq
CLIP-Seq offers the following benefits: (1) high accuracy, faithfully indicating the interaction among molecules in vivo; (2) strong specificity: ultraviolet radiation does not cause protein-protein cross-linking, allowing researchers to directly identify the interaction between RBPs and RNA; and (3) broad application range: particularly suitable for research on splicing factors, RNA binding proteins, miRNA targets, and their interactions.
Workflow and Bioinformatics Analysis Pipeline of CLIP-Seq
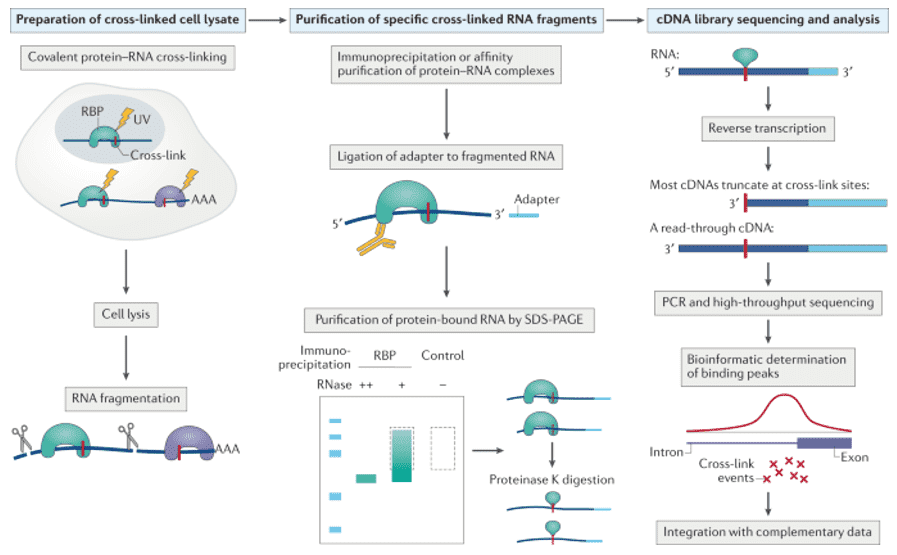
CLIP techniques are more widely used and rely on irradiating cells with UV light, which causes proteins in close proximity to the irradiated bases to form an irreversible covalent bond with the RNA5. The covalent cross-links allow for a thorough purification of the RNA–protein complexes, which is then followed by a series of steps to determine a specific protein's interactions across the transcriptome. CLIP isolates RBP-occupied RNA fragments from cross-linked RNPs using a limited RNase treatment, and sequencing of these fragments can identify RBP binding sites, allowing inference of RBP function by determining the location of binding sites relative to other RBP binding sites or cis-acting elements, for example. CLIP-Seq workflow is depicted in Figure 1.
 Figure 1. General workflow of CLIP-Sequencing (Hafner, 2021)
Figure 1. General workflow of CLIP-Sequencing (Hafner, 2021)
CLIP-Seq assessment involves: (1) sequencing quality distribution, (2) peak calling and visualization, (3) peaks width and distance analysis, (4) defining possible miRNA target(s), (5) distribution analysis of peak, (6) evaluate microRNA-mRNA interactions, (7) protein binding site identification, (8) differential binding analysis, (9) motif search of enrichment sites, and (1) GO and KOG analysis.
Applications of CLIP-Seq
CLIP-seq can be used to confirm the direct interaction of RNA with the target protein as well as the exact binding sites. It can also be used to identify RNA-RBP interaction networks across the entire genome. CLIP-seq can also be used to identify and study the functional mechanisms of lncRNA, circRNA, and miRNA, among other things.
References:
- Hafner M, Katsantoni M, Köster T, et al. CLIP and complementary methods. Nature Reviews Methods Primers. 2021 Mar 4;1(1).
- Bieniasz PD, Kutluay SB. CLIP-related methodologies and their application to retrovirology. Retrovirology. 2018 Dec;15(1).
Read More: